publications
publications
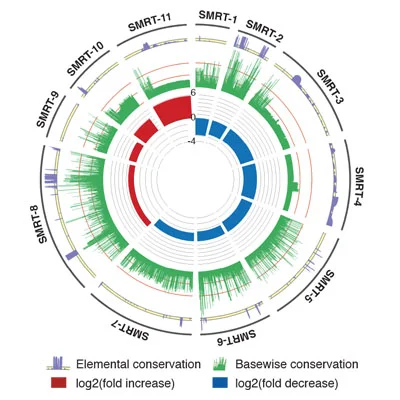
Cancer-Associated Long Noncoding RNA SMRT-2 Controls Epidermal Differentiation.
Lee CS, Mah A, Aros CJ, Lopez-Pajares V, Bhaduri A, Webster DE, Kretz M, Khavari PA. JOURNAL OF INVESTIGATIVE DERMATOLOGY (2018).
Efforts to characterize key genetic anomalies in cutaneous squamous cell carcinoma (SCC) have traditionally focused on changes that affect protein-coding genes, yet altered expression of long noncoding RNAs (lncRNAs) may also play a role in this malignancy. Here we used RNA sequencing to identify novel SCC misregulated transcripts (SMRTs) with features of lncRNAs including SMRT-2, a Ras-regulated transcript altered in SCC that controls epidermal differentiation.
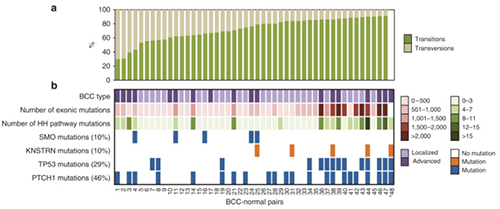
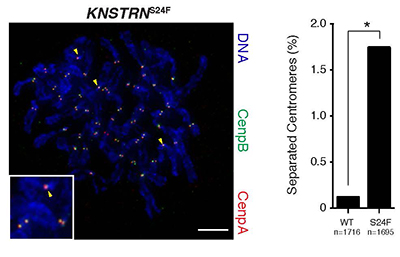
Mutations in the kinetochore gene KNSTRN in basal cell carcinoma.
Jaju PD, Nguyen CB, Mah AM, Atwood SX, Li J, Zia A, Chang AL, Oro AE, Tang JY, Lee CS, Sarin KY. JOURNAL OF INVESTIGATIVE DERMATOLOGY (2015).
Basal cell carcinomas (BCC) are the most common human malignancy. Here we used high throughput exome sequencing to reveal mutations in the recently identified oncogene KNSTRN. As in other ultraviolet light (UV)-associated malignancies, the KNSTRN S24F hotspot mutation results in genomic instability that may enable tumor progression.
Genomic analysis of mycosis fungoides and Sézary syndrome identifies recurrent alterations in TNFR2.
Ungewickell A, Bhaduri A, Rios E, Reuter J, Lee CS, Mah A, Zehnder A, Ohgami R, Kulkarni S, Armstrong R, Weng WK, Gratzinger D, Tavallaee M, Rook A, Snyder M, Kim Y & Khavari PA. NATURE GENETICS (2015).
Mycosis fungoides and Sézary syndrome comprise the majority of cutaneous T cell lymphomas (CTCL) and are known for their clinical heterogeneity as well as limited treatment options. Here we used high throughput sequencing to better characterize the genetic basis of these malignancies and identified recurrent alterations in TNFR2 that regulate T-cell survival and proliferation.
Recurrent point mutations in the kinetochore gene KNSTRN in cutaneous squamous cell carcinoma.
Lee CS, Bhaduri A, Mah A, Johnson WL, Ungewickell A, Aros CJ, Nguyen CB, Rios EJ, Siprashvili Z, Straight A, Kim J, Aasi SZ & Khavari PA. NATURE GENETICS (2014).
Epithelial neoplasms comprise ~90% of human cancers and are characterized by widespread genome damage occurring through mechanisms that are incompletely understood. Here we used high throughput exome sequencing to define recurrent mutations in epidermal squamous cell carcinoma (SCC), the second most common cancer in humans. SCC displays hotspot mutations in the KNSTRN gene that trigger aneuploidy and accelerate tumorigenesis, identifying a new mechanism of genomic injury in cancer.
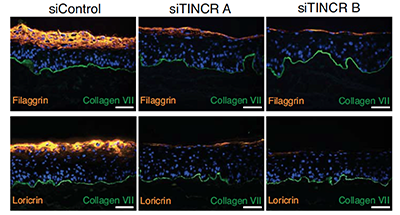
Control of somatic tissue differentiation by the long non-coding RNA TINCR.
Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff A, Chow J, Johnston D, Kim G, Spitale RC, Flynn RA, Zheng G, Aiyer S, Raj A, Rinn JL, Chang HY & Khavari PA. NATURE (2013).
Long noncoding RNAs (lncRNAs) act by a host of mechanisms to impact genomic expression. Here we identified a novel lncRNA named terminal differentiation-induced noncoding RNA (TINCR), which controls expression of hundreds of genes by a novel post-transcriptional mechanism involving complementary base pairing of TINCR to target mRNAs through a 25nt “TINCR box” sequence. TINCR stabilizes bound differentiation gene mRNAs in concert with the Staufen 1 protein, demonstrating an entirely new mechanism for lncRNA action.
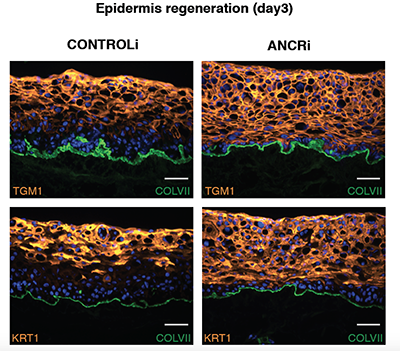
Suppression of progenitor differentiation requires the long noncoding RNA ANCR.
Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE, Rinn JL, Chang HY, Siprashvili Z & Khavari PA. GENES AND DEVELOPMENT (2012).
Long noncoding RNAs (lncRNAs) regulate diverse processes, yet a potential role for lncRNAs in regulating the transition between the progenitor state and a terminally differentiated one remains uncharacterized. Here we used transcriptome sequencing and custom tiling arrays to identify a novel lncRNA named anti-differentiation noncoding RNA (ANCR) that is required to enforce the undifferentiated cell state within the epidermis, demonstrating an entirely new lncRNA function.
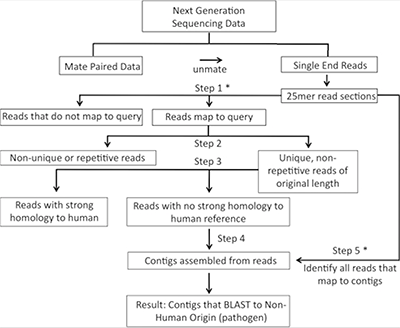
Rapid identification of non-human sequences in high-throughput sequencing datasets.
Bhaduri A, Qu K, Lee CS, Ungewickell A & Khavari PA. BIOINFORMATICS (2012).
The discovery that Merkel cell polyomavirus causes the majority of Merkel cell carcinomas, a rare but aggressive type of skin cancer, illustrates that sequencing-based approaches can successfully identify disease-causing pathogens such as oncoviruses. Here we developed an intersection-based pathogen detection workflow named Rapid Identification of Non-human Sequences (RINS) that employs a user-provided reference set to identify non-human sequences in deep sequencing datasets. RINS identifies non-human sequences with high sensitivity as well as specificity and performs at improved speed and lower cost compared to other available algorithms.
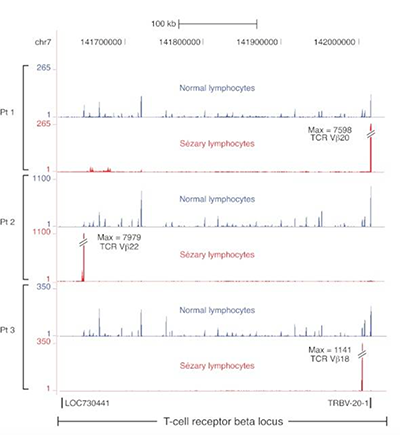
Transcriptome sequencing in Sezary syndrome identifies Sezary cell and mycosis fungoides-associated lncRNAs and novel transcripts.
Lee CS, Ungewickell A, Bhaduri A, Qu K, Webster DE, Armstrong R, Weng WK, Aros CJ, Mah A, Chen RO, Lin M, Sundram U, Chang HY, Kretz M, Kim YH & Khavari PA. BLOOD (2012).
Sézary syndrome (SS) is an aggressive cutaneous T-cell lymphoma (CTCL) of unknown etiology in which malignant cells circulate in the peripheral blood. Here we performed RNA sequencing on malignant Sézary cells as well as patient-matched nonmalignant CD4+ T cells to characterize the SS transcriptome and identify differentially expressed unannotated transcripts with low protein-coding potential that implicate long noncoding RNA (lncRNA) dysregulation in this malignancy.