


Overview
Overview
We are interested in discovering and functionally characterizing new oncogenes and tumor suppressor genes in skin cancer. We seek to understand how these novel genes maintain homeostasis in epithelial tissues in general as well as the skin in particular. We use genomics and proteomics-based approaches combined with animal and human skin tissue models. The ultimate goal of our research is to improve current treatment paradigms and identify actionable therapeutic targets.

Skin Cancer
Skin Cancer
Cancers that arise in epithelial tissue, such as lung, breast, colon, prostate, and skin, account for approximately 90 percent of all human malignancies. These carcinomas affect over 30 million individuals worldwide and are the second leading cause of death in the United States. Work in our lab utilizes high-throughput sequencing approaches to gain a deeper understanding of the diversity, function, and clinical significance of the somatic alterations present in skin cancer genomes. Sequencing of tumors and matched normal tissue has the potential to highlight previously unexplored disease mechanisms, identify prognostic markers, and even transform clinical practice with effective targeted therapies. Insight gained from studying skin cancer may inform our understanding of other epithelial malignancies as well.
We were the first to identify hotspot mutations in the kinetochore gene KNSTRN in cutaneous squamous cell carcinoma (SCC) that trigger aneuploidy and accelerate tumorigenesis [Nature Genetics (2014)].
We also helped demonstrate KNSTRN mutations in basal cell carcinoma (BCC) [Journal of Investigative Dermatology (2015)].
Discovery of the mechanisms that underlie KNSTRN’s pro-tumorigenic effects along with identifying new cancer genes is a major focus of our lab. Work by our group has also focused on cutaneous T-cell lymphoma (CTCL), a non-Hodgkin lymphoma of clonally-derived, skin-homing T-cells that includes mycosis fungoides and Sézary syndrome.
We were the first to define the Sézary cell transcriptome by RNA sequencing [Blood (2012)] and helped establish recurrent alterations in TNFR2 in CTCL that regulate T-cell proliferation and survival using exome sequencing [Nature Genetics (2015)]. These efforts also identified long noncoding RNAs (lncRNAs) that are dysregulated in CTCL with potential roles in this malignancy.

Skin Homeostasis
Skin Homeostasis
Disruptions in epidermal differentiation underlie more than 100 genetic skin conditions with varied clinical and histopathological features. Our group is interested in discovering new protein-coding and noncoding regulators of skin homeostasis and establishing their epistatic relationships within key signaling networks.
We also identified previously uncharacterized skin cancer-associated lncRNAs and unannotated transcripts with features of lncRNAs including SCC Misregulated Transcript (SMRT)-2, a Ras-regulated transcript altered in SCC that mediates epidermal homeostasis [Journal of Investigative Dermatology (2018)]. We helped identify the first lncRNAs involved in somatic stem cell differentiation, including the ANCR stem cell lncRNA [Genes & Development (2012)] and the terminal differentiation lncRNA, TINCR [Nature (2013)]. Given the reciprocal relationship between homeostasis and tumorigenesis, discovery of previously unappreciated mechanisms of cancer progression through a deeper understanding of essential homeostatic regulators is another major focus of our lab.

Models
Models
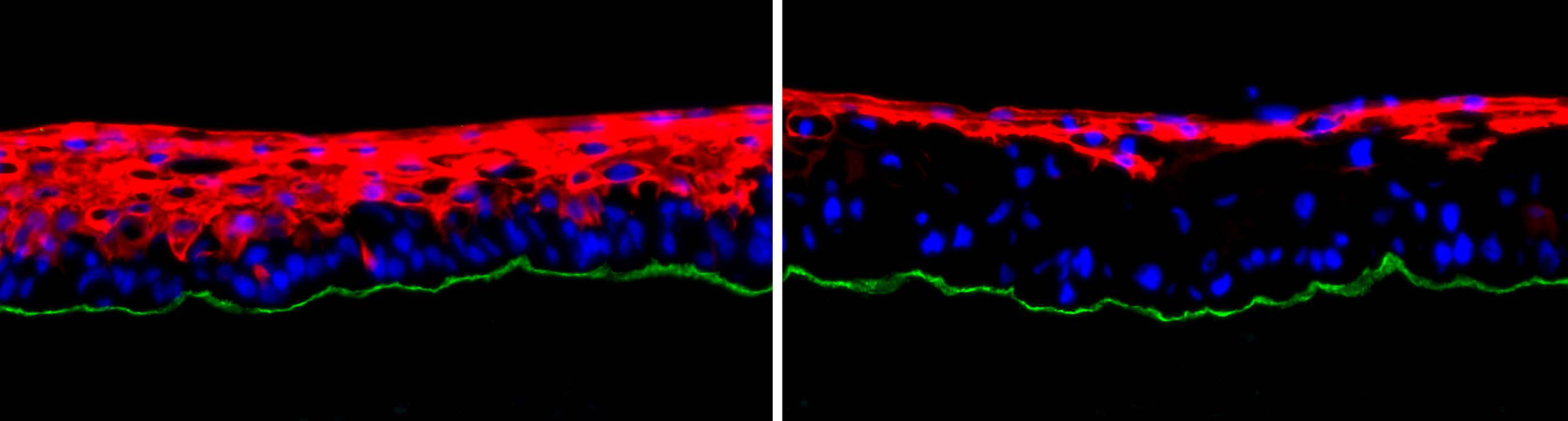
We use human skin tissue models that provide a tractable and clinically relevant platform to study epithelial homeostasis as well as tumor progression. Regenerated human skin tissue comprised of primary human keratinocytes within a 3-dimensional intact architecture that includes the basement membrane and human dermal stroma faithfully recapitulates the global gene expression and architecture of actual human skin. Co-expression of oncogenic Ras with Cdk4 to promote G1 escape in this model rapidly converts normal epidermal tissue into invasive neoplasia that reprises cardinal features of human skin cancer and can be placed as a human tissue xenograft on immune-deficient mice for longer studies. Genetic constructs encoding elements of unknown function can be superimposed into these models to assess their effect on homeostasis and tumor progression by highly quantitative metrics. Multiple (>15) alleles can be altered simultaneously within intact tissue, enabling genetic studies to be performed with a degree of complexity that is often challenging with traditional animal models. Genome editing through a combination of CRISPR/Cas9 and recombinogenic adeno-associated virus (AAV) vectors further allows us to introduce cancer-associated mutations in the endogenous alleles of primary cells. These organotypic human skin models enable human disease to be studied in human tissue, which is known to differ significantly in architecture, metabolism, signal transduction, and DNA repair from mouse and is also differentially resistant to carcinogenesis. These models also incorporate key elements of epithelial tumor progression lacking in subcutaneous injection models, including pre-malignant hyperplasia, in situ neoplasia, and invasion of neoplastic cells across the basement membrane into the surrounding stroma.